import networkx as nx
import EoN
from collections import defaultdict
import matplotlib.pyplot as plt
import scipy
import random
colors = ['#5AB3E6','#FF2000','#009A80','#E69A00', '#CD9AB3', '#0073B3','#F0E442']
rho = 0.01
Nbig=500000
Nsmall = 5000
tau =0.4
gamma = 1.
def poisson():
return scipy.random.poisson(5)
def PsiPoisson(x):
return scipy.exp(-5*(1-x))
def DPsiPoisson(x):
return 5*scipy.exp(-5*(1-x))
bimodalPk = {8:0.5, 2:0.5}
def PsiBimodal(x):
return (x**8 +x**2)/2.
def DPsiBimodal(x):
return(8*x**7 + 2*x)/2.
def homogeneous():
return 5
def PsiHomogeneous(x):
return x**5
def DPsiHomogeneous(x):
return 5*x**4
PlPk = {}
exponent = 1.418184432
kave = 0
for k in range(1,81):
PlPk[k]=k**(-exponent)*scipy.exp(-k*1./40)
kave += k*PlPk[k]
normfact= sum(PlPk.values())
for k in PlPk:
PlPk[k] /= normfact
#def trunc_pow_law():
# r = random.random()
# for k in PlPk:
# r -= PlPk[k]
# if r<0:
# return k
def PsiPowLaw(x):
#print PlPk
rval = 0
for k in PlPk:
rval += PlPk[k]*x**k
return rval
def DPsiPowLaw(x):
rval = 0
for k in PlPk:
rval += k*PlPk[k]*x**(k-1)
return rval
def get_G(N, Pk):
while True:
ks = []
for ctr in range(N):
r = random.random()
for k in Pk:
if r<Pk[k]:
break
else:
r-= Pk[k]
ks.append(k)
if sum(ks)%2==0:
break
G = nx.configuration_model(ks)
return G
report_times = scipy.linspace(0,20,41)
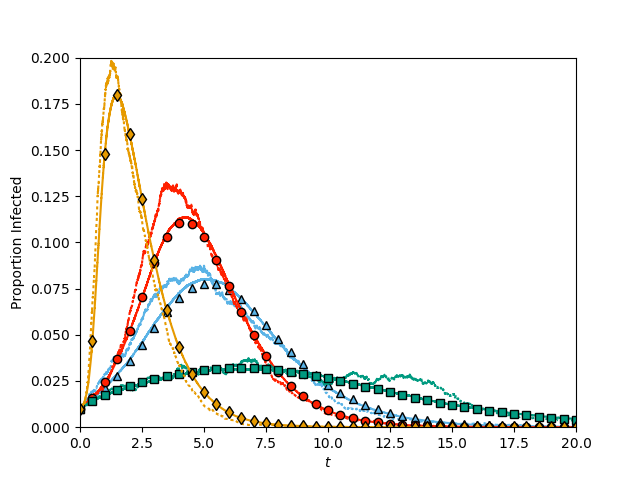
def process_degree_distribution(Gbig, Gsmall, color, Psi, DPsi, symbol):
t, S, I, R = EoN.fast_SIR(Gsmall, tau, gamma, rho=rho)
plt.plot(t, I*1./Gsmall.order(), ':', color = color)
t, S, I, R = EoN.fast_SIR(Gbig, tau, gamma, rho=rho)
plt.plot(t, I*1./Gbig.order(), color = color)
N= Gbig.order()#N is arbitrary, but included because our implementation of EBCM assumes N is given.
t, S, I, R = EoN.EBCM(N, lambda x: (1-rho)*Psi(x), lambda x: (1-rho)*DPsi(x), tau, gamma, 1-rho)
I = EoN.subsample(report_times, t, I)
plt.plot(report_times, I/N, symbol, color = color, markeredgecolor='k')
#Erdos Renyi
Gsmall = nx.fast_gnp_random_graph(Nsmall, 5./(Nsmall-1))
Gbig = nx.fast_gnp_random_graph(Nbig, 5./(Nbig-1))
process_degree_distribution(Gbig, Gsmall, colors[0], PsiPoisson, DPsiPoisson, '^')
#Bimodal
Gsmall = get_G(Nsmall, bimodalPk)
Gbig = get_G(Nbig, bimodalPk)
process_degree_distribution(Gbig, Gsmall, colors[1], PsiBimodal, DPsiBimodal, 'o')
#Homogeneous
Gsmall = get_G(Nsmall, {5:1.})
Gbig = get_G(Nbig, {5:1.})
process_degree_distribution(Gbig, Gsmall, colors[2], PsiHomogeneous, DPsiHomogeneous, 's')
#Powerlaw
Gsmall = get_G(Nsmall, PlPk)
Gbig = get_G(Nbig, PlPk)
process_degree_distribution(Gbig, Gsmall, colors[3], PsiPowLaw, DPsiPowLaw, 'd')
plt.axis(xmin=0, ymin=0, xmax = 20, ymax = 0.2)
plt.xlabel('$t$')
plt.ylabel('Proportion Infected')
plt.savefig('fig6p24.png')