import EoN
import networkx as nx
from matplotlib import rc
import matplotlib.pylab as plt
import scipy
import random
colors = ['#5AB3E6','#FF2000','#009A80','#E69A00', '#CD9AB3', '#0073B3',
'#F0E442']
#commands to make legend be in LaTeX font
#rc('font', **{'family': 'serif', 'serif': ['Computer Modern']})
rc('text', usetex=True)
rho = 0.025
target_k = 6
N=10000
tau = 0.5
gamma = 1.
ts = scipy.arange(0,40,0.05)
count = 50 #number of simulations to run for each
def generate_network(Pk, N, ntries = 100):
r'''Generates an N-node random network whose degree distribution is given by Pk'''
counter = 0
while counter< ntries:
counter += 1
ks = []
for ctr in range(N):
ks.append(Pk())
if sum(ks)%2 == 0:
break
if sum(ks)%2 ==1:
raise EoN.EoNError("cannot generate even degree sum")
G = nx.configuration_model(ks)
return G
#An erdos-renyi network has a Poisson degree distribution.
def PkPoisson():
return scipy.random.poisson(target_k)
def PsiPoisson(x):
return scipy.exp(-target_k*(1-x))
def DPsiPoisson(x):
return target_k*scipy.exp(-target_k*(1-x))
#a regular (homogeneous) network has a simple generating function.
def PkHomogeneous():
return target_k
def PsiHomogeneous(x):
return x**target_k
def DPsiHomogeneous(x):
return target_k*x**(target_k-1)
#The following 30 - 40 lines or so are devoted to defining the degree distribution
#and the generating function of the truncated power law network.
#defining the power law degree distribution here:
assert(target_k==6) #if you've changed target_k, then you'll
#want to update the range 1..61 and/or
#the exponent 1.5.
PlPk = {}
exponent = 1.5
kave = 0
for k in range(1,61):
PlPk[k]=k**(-exponent)
kave += k*PlPk[k]
normfactor= sum(PlPk.values())
for k in PlPk:
PlPk[k] /= normfactor
def PkPowLaw():
r = random.random()
for k in PlPk:
r -= PlPk[k]
if r<0:
return k
def PsiPowLaw(x):
#print PlPk
rval = 0
for k in PlPk:
rval += PlPk[k]*x**k
return rval
def DPsiPowLaw(x):
rval = 0
for k in PlPk:
rval += k*PlPk[k]*x**(k-1)
return rval
#End of power law network properties.
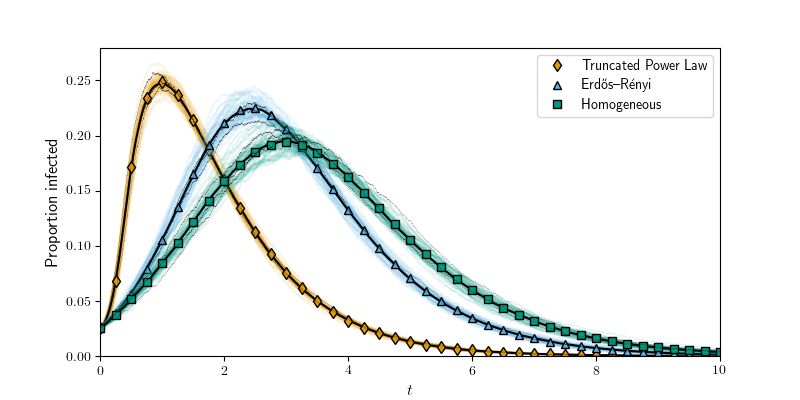
def process_degree_distribution(N, Pk, color, Psi, DPsi, symbol, label, count):
report_times = scipy.linspace(0,30,3000)
sums = 0*report_times
for cnt in range(count):
G = generate_network(Pk, N)
t, S, I, R = EoN.fast_SIR(G, tau, gamma, rho=rho)
plt.plot(t, I*1./N, '-', color = color,
alpha = 0.1, linewidth=1)
subsampled_I = EoN.subsample(report_times, t, I)
sums += subsampled_I*1./N
ave = sums/count
plt.plot(report_times, ave, color = 'k')
#Do EBCM
N= G.order()#N is arbitrary, but included because our implementation of EBCM assumes N is given.
t, S, I, R = EoN.EBCM_uniform_introduction(N, Psi, DPsi, tau, gamma, rho, tmin=0, tmax=10, tcount = 41)
plt.plot(t, I/N, symbol, color = color, markeredgecolor='k', label=label)
for cnt in range(3): #do 3 highlighted simulations
G = generate_network(Pk, N)
t, S, I, R = EoN.fast_SIR(G, tau, gamma, rho=rho)
plt.plot(t, I*1./N, '-', color = 'k', linewidth=0.1)
plt.figure(figsize=(8,4))
#Powerlaw
process_degree_distribution(N, PkPowLaw, colors[3], PsiPowLaw, DPsiPowLaw, 'd', r'Truncated Power Law', count)
#Poisson
process_degree_distribution(N, PkPoisson, colors[0], PsiPoisson, DPsiPoisson, '^', r'Erd\H{o}s--R\'{e}nyi', count)
#Homogeneous
process_degree_distribution(N, PkHomogeneous, colors[2], PsiHomogeneous, DPsiHomogeneous, 's', r'Homogeneous', count)
plt.xlabel(r'$t$', fontsize=12)
plt.ylabel(r'Proportion infected', fontsize=12)
plt.legend(loc = 'upper right', numpoints = 1)
plt.axis(xmax=10, xmin=0, ymin=0)
plt.savefig('fig1p2.pdf')